The NIH Genotype-Tissue Expression (GTEX) project was created to establish a sample and data resource for studies on the relationship between genetic variation and gene expression in multiple human tissues. This track hub shows RNA-seq read coverage signal graphs of GTEx samples in the GTEx midpoint milestone data release (V6, October 2015). A total of 7572 tracks representing samples from 570 adult post-mortem individuals are included.
The individual sample tracks are organized by gender (Female Donors, Male Donors) and age, in 6 tracks, where samples can be selected from a donor vs. tissue matrix.
The Illumina TruSeq protocol was used to create an unstranded polyA+ library sequenced on the Illumina HiSeq 2000 platform to produce 76-bp paired end reads at a depth averaging 50M aligned reads per sample.
Sequence reads for this track were aligned to the hg38/GRCh38 human genome using STAR2 assisted by the GENCODE v24 transcriptome definition. The read coverage signal graphs were produced using the --outWig option of STAR2. Read mapping was performed at UCSC by the Computational Genomics lab, using the Toil pipeline. The resulting bedGraph files were converted to bigWig format for display. The bedGraph files were coordinate converted to the hg19/GRCh37 assembly via the UCSC liftOver tool.
The scientific goal of the GTEx project required that the donors and their biospecimen present with no evidence of disease. The tissue types collected were chosen based on their clinical significance, logistical feasibility and their relevance to the scientific goal of the project and the research community. Postmortem samples were collected from non-diseased donors with ages ranging from 20 to 79. 34.4% of donors were female and 65.6% male.

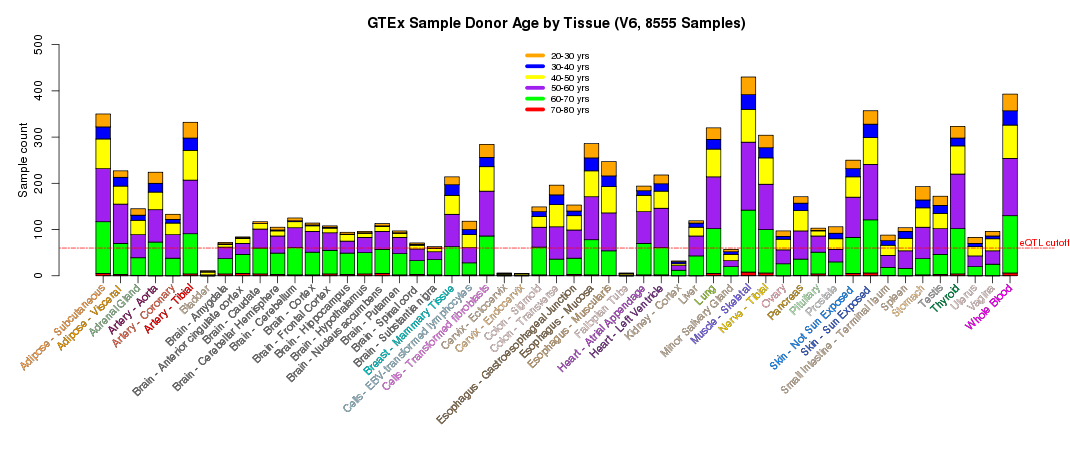
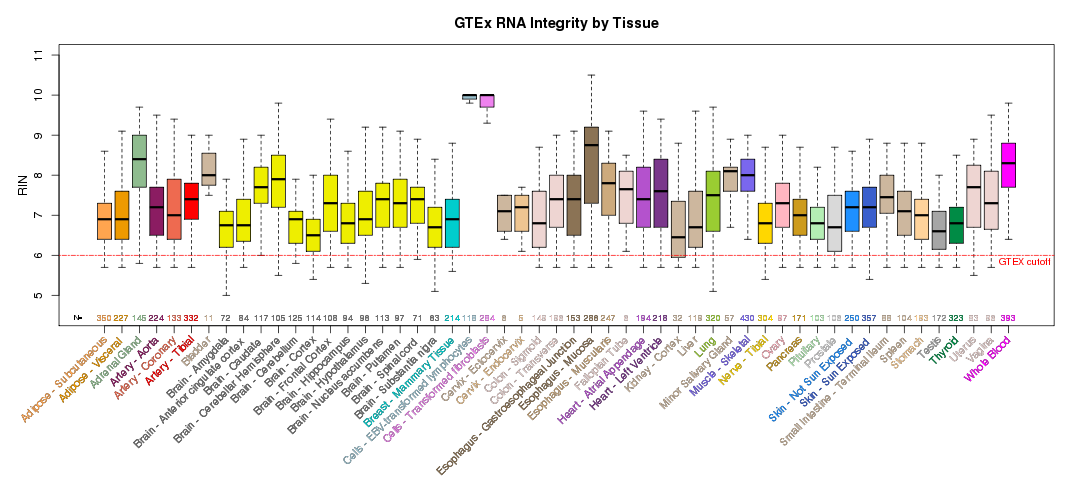
Additional summary plots of GTEx sample characteristics are available at the GTEx Portal Tissue Summary page.
Samples were collected by the GTEx Consortium. RNA-seq was performed by the GTEx Laboratory, Data Analysis and Coordinating Center (LDACC) at the Broad Institute. John Vivian, Melissa Cline, and Benedict Paten of the UCSC Computational Genomics lab were responsible for the sequence read mapping and signal file generation used to produce this hub. Kate Rosenbloom and Parisa Nejad of the UCSC Genome Browser group were responsible for data file post-processing and track hub configuration.
For questions about the GTEx data, contact the GTEx LDACC at the Broad Institute. For questions about this track hub, contact the UCSC Genome Browser mailing list.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013 Jun;45(6):580-5. PMID: 23715323; PMC: PMC4010069
Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv Biobank. 2015 Oct;13(5):311-9. PMID: 26484571; PMC: PMC4675181
Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015 May 8;348(6235):660-5. PMID: 25954002; PMC: PMC4547472DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012 Jun 1;28(11):1530-2. PMID: 22539670; PMC: PMC3356847